An Introduction to Focal Molography
The previous article delved into the world of signal processing, where the challenges we face with environmental noise were underlined. The possibility of acquiring and subsequently processing data in Fourier space was discussed as a potential solution to this problem. But has any such technology ever been developed? In this article, we describe one that has: focal molography.
The Why & The How
As evidenced by our previous articles, the core aim of biosensors is to detect molecular interactions in their natural milieu; this is a challenging task, as the targets we wish to detect are often extremely discrete in environments populated by many other molecules. Many biosensors have been developed thus far to discriminate target molecules from their less relevant counterparts; however, to our knowledge, no sensor has been capable of doing so without also detecting the environmental noise surrounding the molecules of interest. Recently, a new technology has successfully overcome this challenge, allowing it to solely sense molecular interactions, without detecting any environmental noise. The secret to this technology is a concept already well-known by most: holograms.
Molecular Holograms for Light Diffraction
Although many would picture a hologram as an arbitrary 3D object or person made of light, a hologram is simply a pattern that scatters light. The key physical phenomenon occurring in holograms is diffraction, whereby light waves “bend” around obstacles. A typical example for this is a beam of light passing through a thin slit, as shown below.
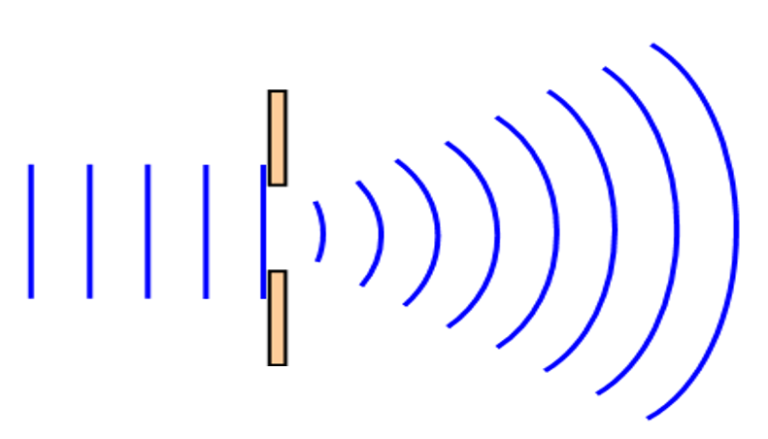
By arranging the obstacles in the path of the light beam in a spatially regular, or “coherent” pattern, the diffraction from the pattern can be precisely tailored in specific directions. Such coherent patterns are commonly used in the form of diffraction gratings, which is none other than a very simple hologram-namely, a pattern on which a deflected light beam is generated. Should these gratings be created using molecules, we can also generate a pattern capable of scattering light; in short, we create a molecular hologram, or a mologram (cf. figure 3a).
Refractometric vs. Diffractometric Biosensors
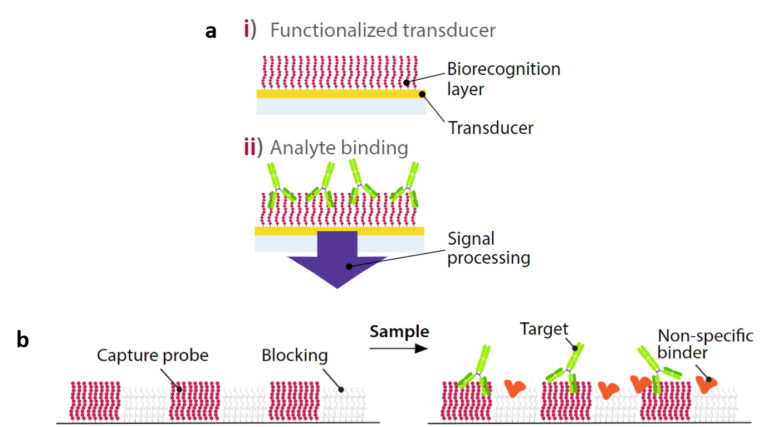
As a reminder, typical biosensors are surface–based devices, where a molecule (capture probe) specific to a target of interest is immobilized onto a polymer coating. This capture probe is in contact with a complex biological sample, containing a vast array of irrelevant molecules. Detecting only the molecules of interest in this sample is the main challenge we are faced with.
As briefly described in the article dedicated to surface plasmon resonance (SPR), many different biosensors utilize the same bio-physical property for molecular detection: the relatively high refractive index of biomolecules compared to water. Here, we distinguish two categories of biosensors: diffractometric (b) and refractometric (a). Both label-free diffractometric and refractometric sensors make use of the high refractive index of biomolecules to detect them, but the key difference between these two groups is how the molecules bind the sensor surface. Indeed, while molecules arbitrarily bind the sensor surface in refractometric sensors, they bind in the form of a molecular hologram in diffractometric sensors. Another important difference is that compared to refractometric biosensors, diffraction-based biosensors are almost completely unresponsive to background signal in the absence of the molecule of interest.
principle of refractometric vs. diffractometric biosensors:
Using Molograms to Filter Noise
Why is it helpful to create molecular patterns, or specifically, molecular gratings? As mentioned above, the light diffraction caused by grating patterns can be tailored. This feature allows us to arrange molecules on the grating, such that the diffracted light is condensed in one single voxel in space.
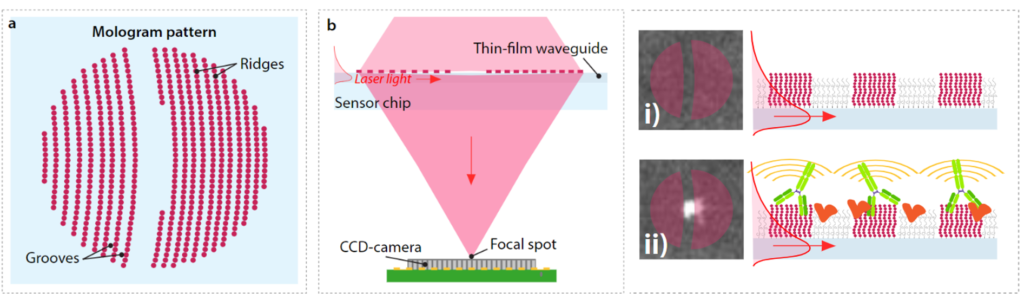
Figure b above depicts this phenomenon: when a laser beam is deflected onto a molecular hologram organized on a grating-type pattern, the light is diffracted. The part of the light beam diffracted by the molecular grating is of particular interest; due to the coherent arrangement of this pattern, the light is also scattered coherently, and is focused in a specific spot that can be measured. So, should this grating be exposed to a biological sample containing an analyte of interest, the target molecules would bind the capture probe only on the ridges. Any other molecule contained in the sample would be found in the grooves; these molecules will also cause light diffraction. However, these light waves will not be diffracted but rather scattered in all spatial directions (i.e. not in the focal point), as the molecules found in the grooves are randomly located. The implications of this are crucial: if we measure signal in the focal spot alone, we solely detect molecular interactions happening on the ridges. In other words, we detect only the molecular interactions of interest.
Still confused? An analogy to put this into perspective.
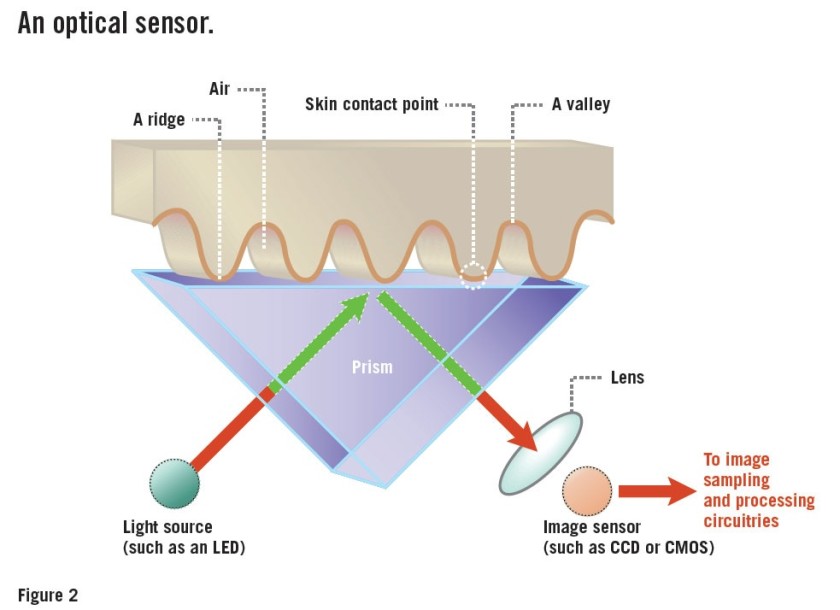
It may still be unclear how molography fits into the signal processing theory discussed in the previous article. Fourier space was described as a superior alternative in the context of data acquisition as it enables the possibility to filter out noise very simply, contrary to noise filtering in real space. How does molography implement this? The mologram forms a diffractive lens that produces a Fourier plane image only a few hundred microns away from the surface of the biosensor. If this is still confusing, here is a simplified way to understand it. Think of a fingerprint reader; when you place your finger onto the scanner and it displays an image of your fingerprint, only the ridges, which contain the relevant information for biometrics, are shown on the image. This is because the grooves do not come into contact
with the imaged surface. One way of seeing this is that the inherent pattern of your finger retains the relevant information and naturally “filters out” the irrelevant information found in the grooves. In short, the pattern itself is a filter! In this respect, molography is the same; by utilizing patterns of molecules, we detect the interactions we are interested in without detecting other analytes.
Outlook
This article introduces a new type of diffractometric biosensor capable of discriminating signal of interest from environmental noise. But what differentiates this technology from other diffractometric biosensors? What other advantages does it possess? How does it compare to other biosensors? The next article will be dedicated to answering these questions!
References
https://sites.google.com/site/wavebehaviourraft/diffraction
https://www.androidauthority.com/how-fingerprint-scanners-work-670934/
Frutiger, A., Fattinger, C. and Vörös, J., 2021. Ultra-Stable Molecular Sensors by Sub-Micron Referencing and Why They Should Be Interrogated by Optical Diffraction—Part I. The Concept of a Spatial Affinity Lock-in Amplifier. Sensors, 21(2), p.469.
Frutiger, A., Gatterdam, K., Blickenstorfer, Y., Reichmuth, A.M., Fattinger, C. and Vörös, J., 2021. Ultra Stable Molecular Sensors by Submicron Referencing and Why They Should Be Interrogated by Optical Diffraction—Part II. Experimental Demonstration. Sensors, 21(1), p.9.
2019 Kübrich Molographic Peptide Arrays Towards Label-free Protein Signaturing in Undiluted Blood Plasma

