Technology
“The reliable detection of molecular interactions in the life sciences might take an unexpected high-tech turn: Lithography on a nanometre scale makes it possible to distinguish between specific and non-specific binding on the surface of a chip.” Dr. Christof Fattinger; 2016
Focal Molography
In short, focal molography is a nanotechnology-based method that cleverly combines photolithography, molecular self-assembly and state-of-the-art optical technology. It is a truly interdisciplinary technology, inspired by physics, tailored for biology and implemented for biomolecular interaction analysis.
Central to the technology is a biological surface structure termed mologram, which is part of our patented sensor chip. Bound biomolecules on the mologram function as “detectives” that recognize the target analyte in the sample through molecular recognition and selective binding and thereby emit a light signal.
This signal indicates the exact measure of binding affinity between recognition site and analyte. This coherent signal is measured in the molographic focus by the instrument’s detector array. One diffraction-limited mologram spot, less than one micrometre in diameter, represents one binding signal. Multiple molograms can be assembled on a tiny chip, meaning that multiple parameters can be measured swiftly, with great selectivity and sensitivity.
This “high-tech turn” was achieved in 2019 and lino Biotech was founded to unleash the potential of diffractometric biosensors for sensitive and robust diffractometric biomolecular interaction analysis.

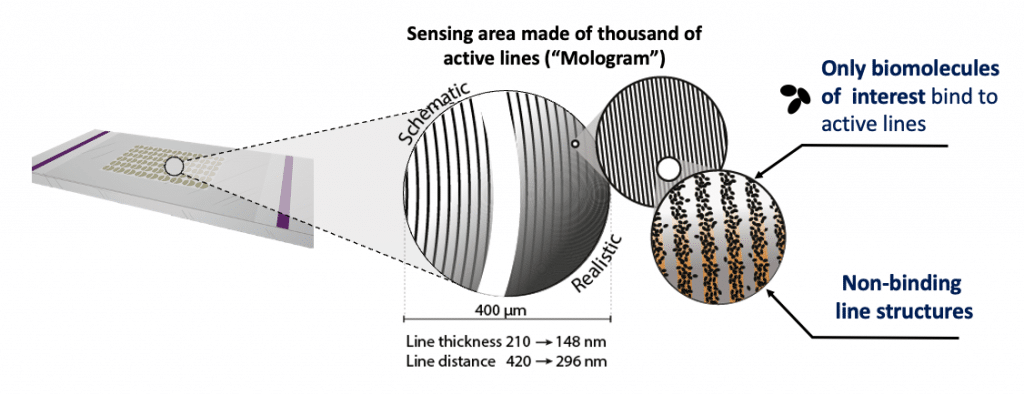
What is a mologram?
A mologram is a biological diffractive element fabricated using biomolecules and nanolithography approaches.
molecules + hologram = mologram
The molgrams are illuminated by a laser beam travelling from left to right across the chip surface. If light encounters biomolecules specifically bound to the active lines on the surface, it is diffracted into a focal point below the chip that can be measured.
Overview
Our core technology provides multidimensional data to characterize biological interactions. Focal Molography is a unique physical principle to measure any kind of interaction. This platform technology opens up new applications due to its six key features. lino Biotech is focusing on the multiplexed characterization in real-time of unlabelled biological material in the bioprocessing sector, where a fast and stable detection in complex media is key.
Focal molography – a new method for Biomolecular Interaction Analysis (BIA)
Biomolecular interaction analysis (BIA), i.e. the direct and label-free monitoring of binding events between molecules on a sensor surface, is a key method in molecular biology. Over the past 30 years, refractometric biosensors, and in particular surface plasmon resonance (SPR), have matured to the de facto standard of BIA despite their significant cross reactivity to environmental and experimental noise sources.
Two publications regarding focal molography introduce and demonstrate the concept of the “spatial affinity lock-in” as a novel design principle to overcome the drawbacks of established BIA methods. The spatial affinity lock-in is analogous to the time-domain lock-in. Instead of a time-domain signal, it modulates the binding signal at a high spatial frequency to separate it from the low spatial frequency environmental noise in Fourier space. Focal molography applies this fundamental detection principle to BIA.
Combined with the right surface chemistry and recognition elements on the sensor surface focal molography enables robust, sensitive and fundamentally new BIA assays such as the direct and label-free monitoring of biomolecular interaction on the cell membrane.
Further reading: Ultra-stable molecular sensors by sub-micron referencing and why they should be interrogated by optical diffraction.
Mologram
at a glance
A biological diffractive element fabricated using biomolecules and nanolithography approaches is called a mologram. The word is a symbiosis from molecules and hologram. The word has its origin in the development of the technology focal molography. It is part of lino Biotech´s sensor chip and functions as detectives that recognize the target analyte in the sample through molecular recognition and selective binding and thereby emit a light signal. A mologram is a coherent assembly of binding sites on a chip that form the blueprint of molecular hologram. The hologram has the shape of a focusing diffractive lens and is illuminated by an evanescent wave. Molecular holograms are diffractometric sensors, which offer an additional mechanism to reduce the effect of nonspecific binding to coherent scattering system. The blueprint of this hologram is encoded into the surface ad-layer. The recognition sites compose a coherent binding pattern, a mologram. Multiple of them can be assembled on a tiny chip by, which means that multiple parameters can be measured swiftly, with great selectivity and sensitivity. In focal molography, the mologram is situated on a high refractive index slab waveguide and illuminated by the fundamental TE mode. Then the affinity modulation is exposed to a biological sample the analyte binds to the molecular hologram, which induces an optical grating that diffracts light from the guided TE mode into a diffraction-limited focal spot.
The stripe pattern you get with focal molography is as described called a mologram. The company has several molograms arranged on a small chip, measuring different molecules on a chip. Bound biomolecules on it function as detectives that recognize the target analyte in the sample through molecular recognition and selective binding and thereby emit a light signal. It is really small with a diameter of 0,4 millimeter and consists of 1,000 very fine stripes. In comparison, the focal point on to which the laser light is focused by it has a diameter of about 1,000th of a millimeter. The possible applications with this technology, being a nanotechnology-based method, are immense and range from real-time measurements to quality control of assay performances.
It is helpful to create molecular patterns as the light diffraction caused by grating patterns can be tailored. This allows lino Biotech to arrange molecules on the grating, such that the diffracted light is condensed in one single voxel in space. When a laser beam is deflected into a mologram organized on a grating-type pattern, the light is diffracted. That part is of particular interest. If the signal in the focal spot alone is measured, it is possible to solely detect molecular interactions happening on the ridges of the mologram. Meaning only detecting the molecular interactions of interest. That technology has a high value for different industries, as measuring multiple parameters in e.g. bioreactors or other pharma measurements for high affinity drug target interactions in real-time over several days are possible, also various measurements for cell and gene therapy manufacturing.
Frequently asked questions
A mologram is a coherent assembly of binding sites on a chip that form the blueprint of the molecular hologram and can also be described as refractive index modulation, caused by target molecules that are bound to a precisely assembled nanopattern of molecular recognition sites.
Mologram is a symbiosis from the word molecules and hologram. It is central to the technology focal molography by lino Biotech and can be described as a biological surface structure., which is part of their patented sensor chip. This coherent nanopattern of molecular recognition sites enables label-free analysis of biochemical interactions in complex samples.
Our Sensor Chips are backfilled, meaning measurements in real complex biological media is possible. This enables a vast range of different assays in a flexible, fast and automated way. We offer different sensor chips for different applications, for example characterizing antibodies with molographic measurements in real complex biological samples with our Protein A Sensor chip is possible.

