Focal Molography, a Guide Inside Cellular Pathways
There are billions of cells in our bodies, each with their own characteristics and functions. To function together, cells need to communicate; during the evolution of multi-cellular organisms, cells have developed communication systems of exceptional complexity. These systems govern every level of communication, and together they dictate cell function, but also tissue function, organ function, and organism function. Understanding these systems is therefore of tremendous value, as issues are often the cause of diseases. In this article, we discuss how Molography could help shed some light on the on-goings of these cellular communication pathways
What are signaling pathways and why are they useful?
Communication between cells occurs mainly via extracellular signaling molecules, i.e. signals that originate from outside the cell. These molecules usually bind a surface receptor on the cell surface, which leads to chain reaction inside the cell, ultimately leading to the corresponding response, which can be a change in gene expression for example, or altered metabolic function within the cell (cf. figure 1). The flow of reactions initiating with a signaling molecule and ending with a cellular response, including all intermediate steps is called a cell signaling pathway.
There are hundreds of molecules that can function as extracellular signals; these molecules then specifically bind a receptor on the target cell. This binding specificity ensures that the target cell responds only to the appropriate signal. The appropriate cellular response is also ensured by the fact that each cell is programmed to respond only to a specific cocktail and specific combinations of signals. This is an extremely elegant solution: with just a small subset of signaling molecules, an almost unlimited number of combinations can be used to relay a signal and control cell behavior. With various cell types responding in different ways to a given signal combination they receive, the complexity can be further increased. Thus, a given signaling molecule often has different effects on different types of cells
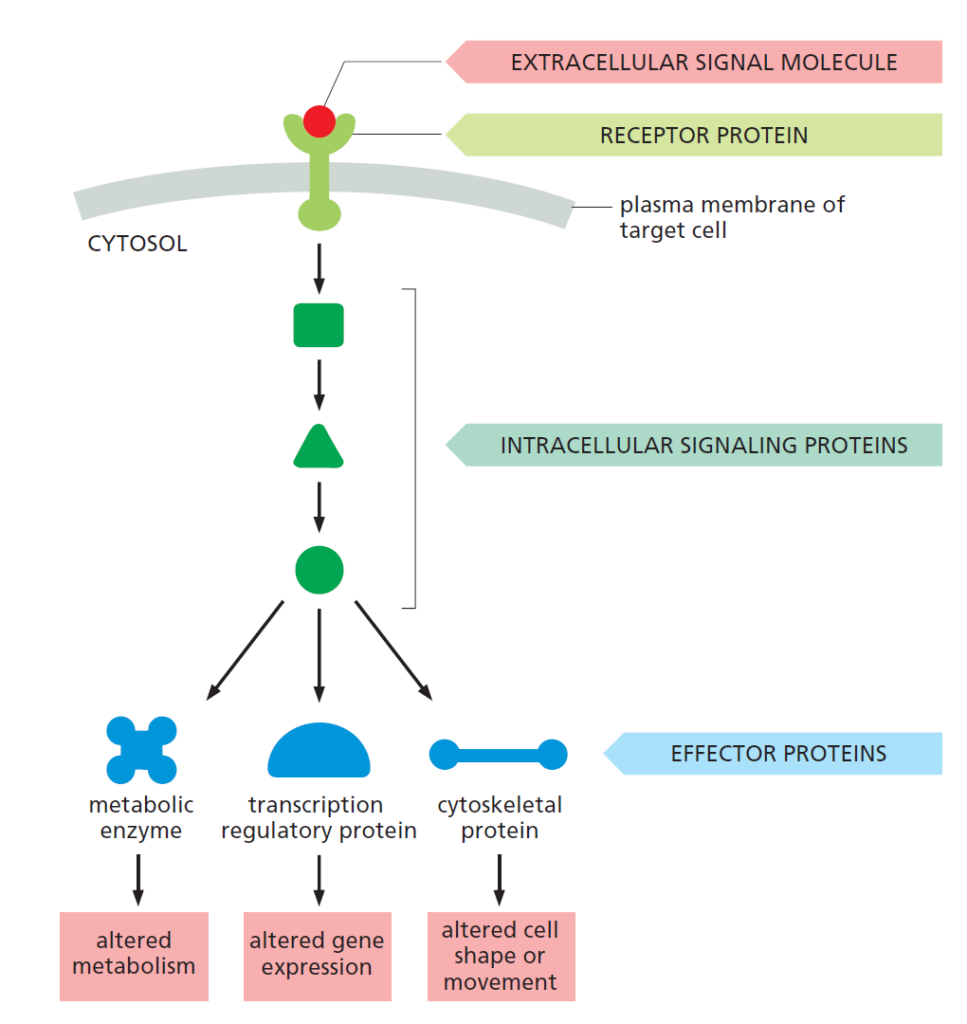
Surface Receptors
Because it is difficult to enter the cell via direct permeation through the cell surface membrane, the majority of signaling molecules bind a surface molecule called a receptor. Receptors are categorized into three broad categories: ion-channel-coupled receptors, G-protein-coupled receptors, and enzyme-coupled receptors. Each type triggers a different subsequent reaction within the cell, and are activated by different kinds of extracellular signaling molecules.
By far the most common receptor type is the second category: G-protein-coupled receptors (GPCRs), that act by regulating the activity of a cell membrane-bound protein (typically an ion-channel or an enzyme) via a molecule called the G protein. The activation of a GPCR pathway can be mediated by hormones, neurotransmitters, and many other extracellular signaling molecules. Many bodily functions rely on GPCR signaling, for example our senses: sight, smell, and taste all depend on them. Indeed, we have over 800 GPCRs dedicated to the sense of smell alone! Because of their vast diversity and their omnipresence in organism function, GPCRs are the most common targets for drug development. Furthermore, their presence on the cell surface makes them excellent targets as they are easily accessible by pharmaceuticals, without requiring the additional challenges of entering the cell.
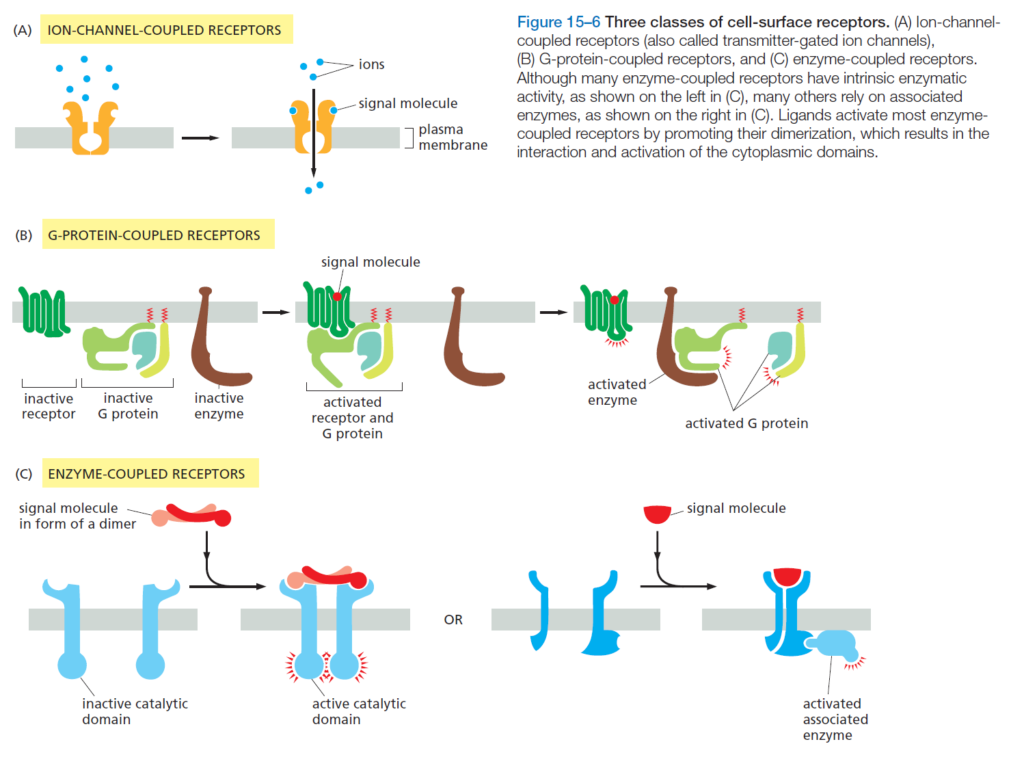
A look inside the cell
Around half of all known drugs to date target GPCRs and their respective signaling pathways. However, despite our growing knowledge on GPCRs and their various functions, many of these remain unknown: “of the many hundreds of genes in the human genome that encode GPCRs, about 150 encode orphan receptors, for which the ligand (the molecule that binds it) is unknown.” These orphan receptors leave a vast space for new drugs that have yet to be discovered, and are a large area of research in the field of drug discovery.
One of the issues faced in drug discovery, particularly related to orphan GPCRs, is that we often only have a view on what is happening outside the cell. One of the simplest ways one can go about researching orphan GPCRs is by exposing a given cell to a chosen signal, selected with an educated guess on what molecule could bind the orphan GPCR of interest, and observe what happens. This method is however extremely ineffective, as it essentially involves looking for a needle in haystack. Furthermore, the chain of events happening inside the cell remains completely unknown: a black box phenomenon. This is particularly problematic in cell signaling as a given signaling molecule can trigger different pathways in different cell types, meaning the end effect can vastly differ amongst cell types.
Traditionally, cell signaling pathways have been analyzed using biochemical methods, as they enable high specificity analysis and are relatively easy to perform. However, as made clear by this article, understanding the individual steps of a signaling pathway, i.e. their mechanism of action, has become increasingly important for applications such as drug development. For this kind of cellular analysis, simple biochemistry is insufficient, and cell-based assays are required; nevertheless, the specificity of the analysis enabled by biochemistry cannot be forgone, and in order to analyze cells with the same degree of molecule specificity, current cell-based assays either heavily rely on labeling techniques and molecular tags to “follow” a given pathway, or utilize label-free technologies which only provide bulk information (i.e. global cellular effects), to the expense of high molecular specificity. What can be done to solve these issues? How can we get better insight on the internal events of a cell without compromising specificity?
Applications
Note: the understanding of this section requires basic knowledge on focal molography. Check out our dedicated blog article here.
The discovery of new drugs targeting orphan GPCRs underscores the need for a cell-based assay capable of analyzing interactions with molecular-level specificity (in this instance, protein-protein interactions), an importantly, without the use of labels, as they likely affect the natural physiology of a given molecular interaction. Focal molography offers an elegant solution to this problem: compatible with whole, living cells, it enables the visualization of molecular interactions occurring at the cell membrane via the spatial ordering of any protein of interest (in this case a GPCR) into a coherent pattern, the mologram. Any molecular interaction happening with these ordered GPCRs become visible and measurable in real-time, while all other interactions, happening outside of the mologram pattern, are not detected. This setup also allows for the analysis of intracellular binding interactions, as the molographic pattern is transferred inside the cell, forming a “transmembrane mologram”. Thus, molecules interacting with the coherently arranged GPCRs, both extracellularly and intracellularly, become visible in the focal spot.
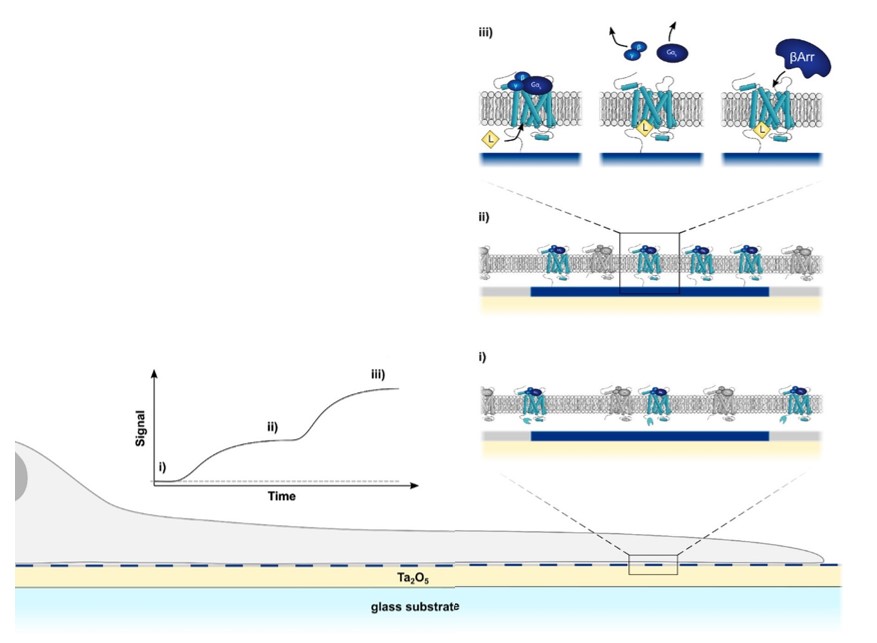
This new way of measuring molecular interactions in live cells is termed “cell-based focal molography”. Its concept goes far beyond GPCRs; indeed, this assay could be used to analyze any membrane bound proteins such as ion channels, immune receptors, and many more. With the features described in this article, focal molography could become a go-to tool for the discovery of new drug targets for all kinds of diseases. For further information on lino’s experimental research and proof-of-concept experiments in this area, you can read one of our published papers on the topic:
Anal. Chem. 2020, 92, 13, 8983–8991
Publication Date: June 11, 2020
https://doi.org/10.1021/acs.analchem.0c00987
References
Molecular Biology of the Cell, Sixth Edition
Anal. Chem. 2020, 92, 13, 8983–8991, 11.06.2020, https://doi.org/10.1021/acs.analchem.0c00987,
https://pubmed. ncbi.nlm.nih.gov/20829029/

